GA4GH Approves Beacon v2
A major step towards federated analysis of biomedical genomics data
![]() At its Spring 2022 GA4GH Connect stakeholder meeting in Montreal (and online...) the
steering committee of the Global Alliance for Genomics and Health (GA4GH)
approved the major "v2" update of the Beacon protocol as official GA4GH standard.
At its Spring 2022 GA4GH Connect stakeholder meeting in Montreal (and online...) the
steering committee of the Global Alliance for Genomics and Health (GA4GH)
approved the major "v2" update of the Beacon protocol as official GA4GH standard.
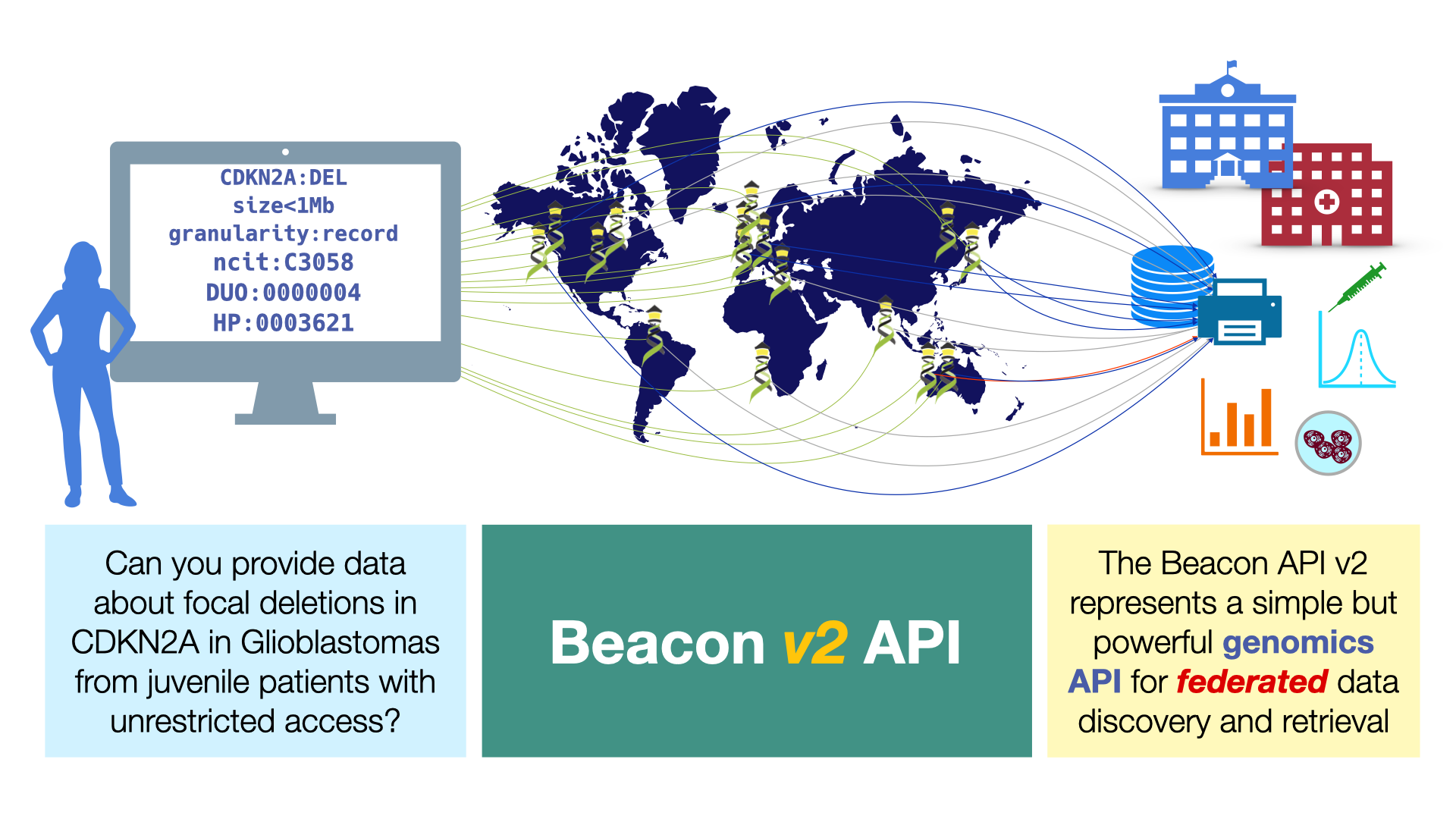
Beacon v2 represents a major revision of the original concept, with a completely new code base and a focus on high flexibility through its modular design. The Beacon code base includes both the framework - i.e. the specific ation for the general formats & components of requests and responses to a Beacon v2 server - as well as the default model which provides schemas for the representation of data from different domains (e.g. genomic variations, biosamples) as well as query parameters.
With respect to the exchange of information about structural genome variation data, Beacon v2 explicitely allows for CNV queries either through simple genomic "Range Queries" as well as "Bracket Queries", for which individual genomic brackets for start and end positions can be specified. Please refer to the documentation for further details.
Also included in the documentation is a comparison of the "variant type" use across different specifications, e.g. how CNVs (and their subtypes) are represented in EFO, VCF, VRS ... and in Beacon queries. This part of the documentation (and the Beacon specification parts) has been heavily influenced by contributions from several members of the hCNV communty. Please watch the space there for updates - and consider to contribute!
Citation
Beacon v2 and Beacon Networks: a "lingua franca" for federated data discovery in biomedical genomics, and beyond. Jordi Rambla, Michael Baudis, Tim Beck, Lauren A. Fromont, Arcadi Navarro, Manuel Rueda, Gary Saunders, Babita Singh, J.Dylan Spalding, Juha Tornroos, Claudia Vasallo, Colin D.Veal, Anthony J.Brookes. Human Mutation (2022) DOI.