ELIXIR Human Copy Number Variation community¶
Among the different types of inherited and acquired genomic variants, regional genomic copy number variations (CNV) contribute - if measured by affected genomic sequences - contribute by far the largest amount of genomic changes, contributing both to many syndromic diseases as well as the vast majority of human cancers. The website of the Human Copy Number Variation Community (hCNV) is a resource originated in ELIXIR's h-CNV Community Implementation Study (2019-2021) with the aim to provide a resource hub and knowledge exchange space for scientists and practitioners working with - or being interested in - genomic copy number variations in health and diseases. However, the scope of the community extends beyond CNVs and includes definition of and work with other types of genomic variations with a focus on structural variants.
CNV News and Announcements¶
The hCNV Community presents itself at ELIXIR Rare Diseases
ELIXIR RD Meeting, Bologna 2025
The ELIXIR hCNV community was a guest at the Rare Disease community meeting in Bologna on May 15, 2025. The hCNV community presented an overview of its activities and several members gave talks on specific topics related to their activities in the hCNV community.
Continue readingA systematic benchmark of copy number variation detection tools for high density SNP genotyping arrays
M.N. van Baardwijk, L.S.E.M. Heijnen, H. Zhao, M. Baudis and A.P. Stubbs¶
Genomics (Elsevier). 2024 Nov 14.¶
- doi: 10.1016/j.ygeno.2024.110962
- PMID: 39547585
 Abstract Copy Number Variations (CNVs) are crucial in various diseases, especially cancer, but detecting them accurately from SNP genotyping arrays remains challenging. Therefore, this study benchmarked five CNV detection tools-PennCNV, QuantiSNP, iPattern, EnsembleCNV, and R-GADA-using SNP array and WGS data from 2002 individuals of the DRAGEN re-analysis of the 1000 Genomes project. Results showed significant variability in tool performance. R-GADA had the highest recall but low precision, while PennCNV was the most reliable in terms of precision and F1 score. EnsembleCNV improved recall by combining multiple callers but increased false positives.
Continue reading
Abstract Copy Number Variations (CNVs) are crucial in various diseases, especially cancer, but detecting them accurately from SNP genotyping arrays remains challenging. Therefore, this study benchmarked five CNV detection tools-PennCNV, QuantiSNP, iPattern, EnsembleCNV, and R-GADA-using SNP array and WGS data from 2002 individuals of the DRAGEN re-analysis of the 1000 Genomes project. Results showed significant variability in tool performance. R-GADA had the highest recall but low precision, while PennCNV was the most reliable in terms of precision and F1 score. EnsembleCNV improved recall by combining multiple callers but increased false positives.
Continue reading
Copy number variation heterogeneity reveals biological inconsistency in hierarchical cancer classifications
Research Article
Ziying Yang, Paula Carrio-Cordo and Michael Baudis¶
Molecular Cytogenetics (Spring Nature). doi: 10.1186/s13039-024-00692-2¶
Abstract: Cancers are heterogeneous diseases with unifying features of abnormal and consuming cell growth, where the deregulation of normal cellular functions is initiated by the accumulation of genomic mutations in cells of - potentially - any organ. At diagnosis malignancies typically present with patterns of somatic genome variants on diverse levels of heterogeneity. Among the different types of genomic alterations, copy number variants (CNV) represent a distinct, near-ubiquitous class of structural variants. Cancer classifications are foundational for patient care and oncology research. Terminologies such as the National Cancer Institute Thesaurus provide large sets of hierarchical cancer classification vocabularies and promote data interoperability and ontology-driven computational analysis. To find out how categorical classifications correspond to genomic observations, we conducted a meta-analysis of inter-sample genomic heterogeneity for classification hierarchies on CNV profiles from 97,142 individual samples across 512 cancer entities, and evaluated recurring CNV signatures across diagnostic subsets. Our results highlight specific biological mechanisms across cancer entities with the potential for improvement of patient stratification and future enhancement of cancer classification systems and provide some indications for cooperative genomic events across distinct clinical entities.
Continue readingCall for Papers BMC Molecular Cytogenetics
Computational Insights into Chromosomal Aberrations: Advancing Molecular Cytogenetics using Genomics Databases
Edited by Jeremy Squire, Federal University of São Paulo, Brazil¶
 With potential interest for members of the CNV communities, BMC Molecular Cytogenetics
is calling for submissions to a new Collection on Computational Insights into
Chromosomal Aberrations: Advancing Molecular Cytogenetics using Genomics Databases.
With potential interest for members of the CNV communities, BMC Molecular Cytogenetics
is calling for submissions to a new Collection on Computational Insights into
Chromosomal Aberrations: Advancing Molecular Cytogenetics using Genomics Databases.
The ELIXIR hCNV Community - Making complex genomics accessible
ELIXIR Webcast
Michael Baudis, Antonio Rausell & Krzysztof Poterlowicz¶
Abstract Genomic copy number variants (CNV) are a major contributor to human genome variation and important factors in rare disease genetics and cancer genomics. However, the complexity of CNV detection technologies, the lack of standardised annotation formats and the fragmentation of cytogenetic and genomic communities so far has limited large scale utilization of CNV profiles in computational genomics. Continue reading
cancercelllines.org - a Novel Resource for Genomic Variants in Cancer Cell Lines
DATABASE Article
Rahel Paloots and Michael Baudis¶
Database (Oxford). 2024 Apr 30:2024:baae030. doi: 10.1093/database/baae030¶
bioarXiv preprint (2023-12-13): https://doi.org/10.1101/2023.12.12.571281¶
 Abstract: Cancer cell lines are an important component in biological and medical research, enabling studies of cellular mechanisms as well as the development and testing of pharmaceuticals. Genomic alterations in cancer cell lines are widely studied as models for oncogenetic events and are represented in a wide range of primary resources. We have created a comprehensive, curated knowledge resource - cancercelllines.org - with the aim to enable easy access to genomic profiling data in cancer cell lines, curated from a variety of resources and integrating both copy number and single nucleotide variants (SNVs) data. We have gathered over 5,600 copy number profiles as well as SNV annotations for 16,000 cell lines and provide this data with mappings to the GRCh38 reference genome. Both genomic variations and associated curated metadata can be queried through the GA4GH Beacon v2 API and a graphical user interface with extensive data retrieval enabled using GA4GH data schemas under a permissive licensing scheme.
Abstract: Cancer cell lines are an important component in biological and medical research, enabling studies of cellular mechanisms as well as the development and testing of pharmaceuticals. Genomic alterations in cancer cell lines are widely studied as models for oncogenetic events and are represented in a wide range of primary resources. We have created a comprehensive, curated knowledge resource - cancercelllines.org - with the aim to enable easy access to genomic profiling data in cancer cell lines, curated from a variety of resources and integrating both copy number and single nucleotide variants (SNVs) data. We have gathered over 5,600 copy number profiles as well as SNV annotations for 16,000 cell lines and provide this data with mappings to the GRCh38 reference genome. Both genomic variations and associated curated metadata can be queried through the GA4GH Beacon v2 API and a graphical user interface with extensive data retrieval enabled using GA4GH data schemas under a permissive licensing scheme.
Availability and Implementation: Our resource is publicly available on the web at cancercelllines.org.
Continue readinglabelSeg: segment annotation for tumor copy number alteration profiles
A tool to assign relative SCNA levels to segments
Hangjia Zhao and Michael Baudis¶
Briefings in Bioinformatics (Oxford). 2024 Jan 31;2024:bbad541.¶
- doi: 10.1093/bib/bbad541
- PMID: 38300514
- bioRxiv. doi: 10.1101/2023.05.17.541097
Abstract Somatic copy number alterations (SCNAs) are a predominant type of oncogenomic alterations that affect a large proportion of the genome in the majority of cancer samples. Current technologies allow high-throughput measurement of such copy number aberrations, generating results consisting of frequently large sets of SCNA segments. However, the automated annotation and integration of such data are particularly challenging because the measured signals reflect biased, relative copy number ratios. In this study, we introduce labelSeg, an algorithm designed for rapid and accurate annotation of CNA segments, with the aim of enhancing the interpretation of tumor SCNA profiles. Continue reading
Beaconize this: Databases for Cancer Genomics and the Development of Open Data Standards
Seminar at the Bioinformatics club of the Centre de Recherche des Cordeliers (CRC)
Université Paris Cité
Michael Baudis¶
In this seminar at the Centre de Recherche des Cordeliers in Paris Michael presents the work of the group, with special emphasis on the role of the Progenetix oncogenomics resources and tools in the development, implementation and testing of the Beacon standard of the Global Alliance for Genomics and Health (GA4GH).
Continue readinghCNV Community Meeting 2023
A visit to the Hinxton motherlode...
 On the last day of November 2023 members of the ELIXIR hCNV community meet on the
Wellcome Trust Genome Campus in Hinxton for a hCNV community event. Main topics
are the definition of achievable short term goals with intersecting interests and
expertise, as well as how to aquire long term support for the community's activities.
On the last day of November 2023 members of the ELIXIR hCNV community meet on the
Wellcome Trust Genome Campus in Hinxton for a hCNV community event. Main topics
are the definition of achievable short term goals with intersecting interests and
expertise, as well as how to aquire long term support for the community's activities.
CNV Project at biohackathon23
Participating at #BioHackEU23 in Barcelona with a CNV reference resource project
 Members of the hCNV community participate at this year's
Biohackathon Europe event. The main project will address the creation of the template
for a "beaconized" public resource for reference (i.e. not disease associated)
copy number variation data, including the necessary tooling for the import from
e.g. VCF or BED file variants into Beacon backends (such as our
Members of the hCNV community participate at this year's
Biohackathon Europe event. The main project will address the creation of the template
for a "beaconized" public resource for reference (i.e. not disease associated)
copy number variation data, including the necessary tooling for the import from
e.g. VCF or BED file variants into Beacon backends (such as our bycon environment).
Progenetix as SIB and ELIXIR Resource
Recognizing the Progenetix platform as Swiss contribution to the European bioinformatics resources ecosystem
 The Progenetix resource has finally been recognized as an official contribution to the ELIXIR European bioinformatics informatics ecosystem. Besides Expasy Progenetix now is linked through ELIXIR's resource page. Or just go directly to progenetix.org (and its daughter project cancercelllines.org).
The Progenetix resource has finally been recognized as an official contribution to the ELIXIR European bioinformatics informatics ecosystem. Besides Expasy Progenetix now is linked through ELIXIR's resource page. Or just go directly to progenetix.org (and its daughter project cancercelllines.org).
CNV Annotation Standards
New page dedicated to the documentation of CNV standards
We have updated an earlier page that linked to different cytogeneic & genomic annotation standards with an extensive table delineating CNV labeling in several file and API formats. The documentation itself is now linked from the main menue for easier access.
Continue readingGA4GH special interest group.
GA4GH special interest group working on Structural Varation Representation
GA4GH is seeking members to participate in new special interest group that would tackle the problem of representing structural variations in sequenced DNA
Continue readingCandidate targets of copy number deletion events across 17 cancer types
Identifying cancer related genes against the background of somatic CNV events
Huang Q and Baudis M¶
doi: 10.3389/fgene.2022.1017657¶
previous bioRxiv (first )2022-06-29), doi.org/10.1101/2022.06.29.498080¶
Abstract Genome variation is the direct cause of cancer and driver of its clonal evolution. While the impact of many point mutations can be evaluated through their modification of individual genomic elements, even a single copy number aberration (CNA) may encompass hundreds of genes and therefore pose challenges to untangle potentially complex functional effects. However, consistent, recurring and disease-specific patterns in the genome-wide CNA landscape imply that particular CNA may promote cancer-type-specific characteristics. Discerning essential cancer-promoting alterations from the inherent co-dependency in CNA would improve the understanding of mechanisms of CNA and provide new insights into cancer biology and potential therapeutic targets. Continue reading
ELIXIR All Hands 2022 - h-CNV Representation
Workshop proposals | Amsterdam 7-10 June 2022
The hCNV community has been invited to participate in 3 workshops:
Continue readinghCNV Implementation Studies Old and New
Michael Baudis @ ELIXIR Human Data Communities
This presentation provided an overview about the hCNV community, implementation studies and ongoing work, e.g. interaction with the GA4GH VRS standard group and Beacon development.
Continue readingCNV Ontology Proposal - Now Live at EFO
EFO Ontology contains now terms for (relative) CNV levels
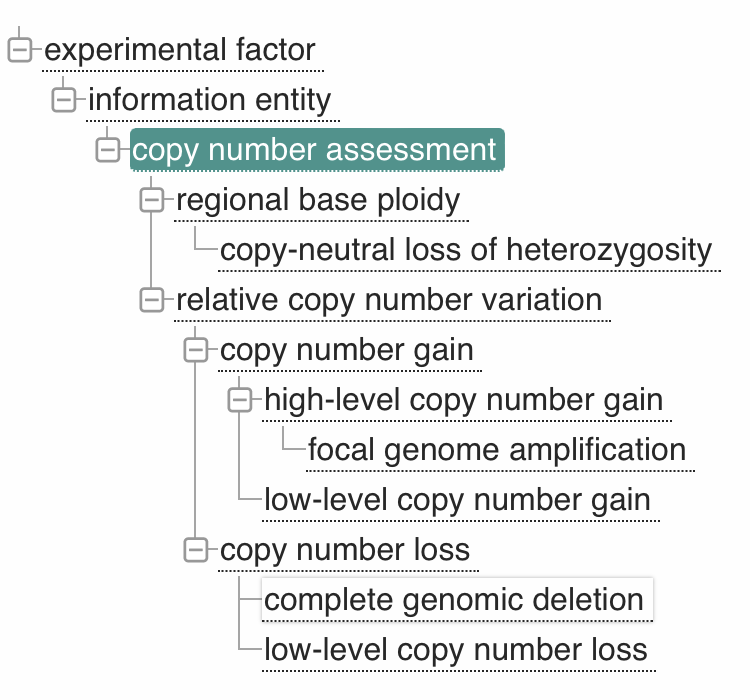 As part of the hCNV-X work - related to "Workflows and Tools for hCNV Data Exchange
Procedures" and to the intersection with Beacon and GA4GH VRS - we have now a new
proposal
for the creation of an ontology for the annotation of (relative) CNV events. The CNV
representation ontology is targeted for adoption by Sequence Ontology (SO)
and then to be used by an updated version of the VRS standard. Please see the
discussions linked from the proposal page.
However, we have also contributed the CNV proposal to EFO where it has gotten
live on January 21.
As part of the hCNV-X work - related to "Workflows and Tools for hCNV Data Exchange
Procedures" and to the intersection with Beacon and GA4GH VRS - we have now a new
proposal
for the creation of an ontology for the annotation of (relative) CNV events. The CNV
representation ontology is targeted for adoption by Sequence Ontology (SO)
and then to be used by an updated version of the VRS standard. Please see the
discussions linked from the proposal page.
However, we have also contributed the CNV proposal to EFO where it has gotten
live on January 21.
hCNV Site now at cnvar.org
The homepage of the hCNV community has now been mapped to the cnvar.org domain.
Continue readingThe Progenetix oncogenomic resource in 2021
An article about current state and recent changes to th eProgenetix resource
Qingyao Huang, Paula Carrio Cordo, Bo Gao, Rahel Paloots, Michael Baudis¶
Database (Oxford). 2021 Jul 17;2021:baab043.¶
- doi: 10.1093/database/baab043.
- PMID: 34272855
- PMCID: PMC8285936.
- bioRxiv. doi: doi.org/10.1101/2021.02.15.428237
 This article provides an overview of recent changes and additions to the Progenetix database and the services provided through the resource.
This article provides an overview of recent changes and additions to the Progenetix database and the services provided through the resource.
hCNV Community and Implementation Studies
Presentation at ELIXIR All Hands 2021
At the ELIXIR All Hands 2021 Human Data Day Michael presents a very brief overview of the ending and upcoming ELIXIR hCNV implementation studies.
Continue readingCNV Beacon at Biohackathon Europe 2020
CNV detection software containerisation and benchmarking
For the Biohackathon Europe (Nov 08-22), the hCNV project participates with a "containerisation" project.
Continue readingPR: The ELIXIR hCNV Community publishes its programme
ELIXIR h-CNV press release
The ELIXIR Community on Human Copy Number Variations (hCNV) has presented its work programme. In a paper published in the ELIXIR F1000R gateway, the Community defines seven strategic objectives to support human CNV detection and interpretation ... (please read on elixir-europe.org)
Continue readingThe ELIXIR hCNV Community
White Paper Published @ F1000Research
The ELIXIR Human Copy Number Variations Community: building bioinformatics infrastructure for research¶
Salgado D, Armean IM, Baudis M et al.¶
F1000Research 2020, 9 (ELIXIR):1229¶
Abstract¶
Copy number variations (CNVs) are major causative contributors both in the genesis of genetic diseases and human neoplasias. While “High-Throughput” sequencing technologies are increasingly becoming the primary choice for genomic screening analysis, their ability to efficiently detect CNVs is still heterogeneous and remains to be developed.
Continue readingHGVS 2019 Gothenburg
CNV Satellite Meeting @ Human Genome Variation Society - Gothenburg 2019
As a satellite meeting of the European Human Genetics Conference, the HGVS2019 meeting this year had a focus on human Copy Number Number Variations.
Continue readingh-CNV kick-off
h-CNV community kick-off meeting at ELIXIR All Hands in Lisbon
The h-CNV community had its kick-off meeting at the ELIXIR All Hands meeting in Lisbon, June 17-20 2019.
The meeting was used to present and discuss the different work packages and overall organisation of the project (slides attached).
Links¶
- presentation slides (PDF)
Launch of the ELIXIR h-CNV Community
A new ELIXIR community
The first h-CNV Community Implementation Study was officially launched on June 1st, 2019.
The project has an initial time span of two years. For further information please follow the link to the ELIXIR project page.
Continue reading